INSIGHTS INTO THE EVOLUTION OF THE CHLOROPLAST GENOME AND THE PHYLOGENY OF BEGONIA
DOI:
https://doi.org/10.24823/ejb.2022.408Keywords:
Begonia, Begoniaceae, Hillebrandia, IR expansion, Plastome, Species barcodeAbstract
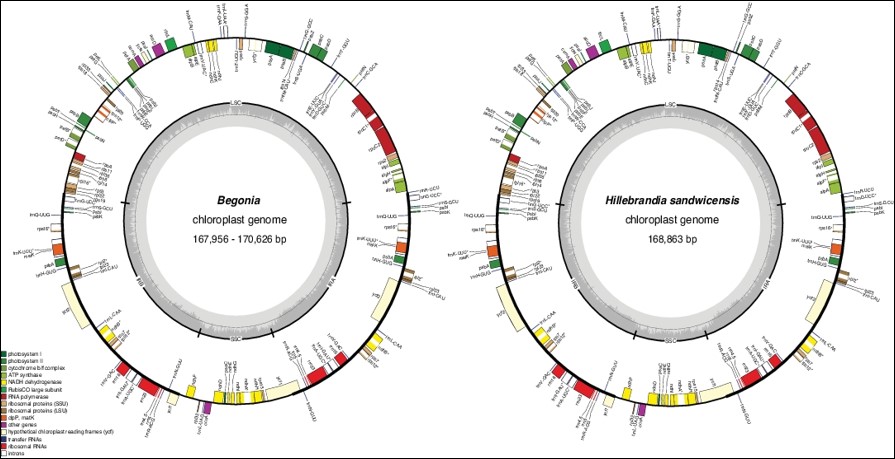
Begonia (Begoniaceae) is one of the largest angiosperm genera, comprising more than 2000 species; this makes it ideal as a model to investigate the genomic basis of species radiations. Here we present the results of the first genus-wide comparative study of plastid genome structure, sequence diversity, and phylogenetics of Begoniaceae, in which 44 complete Begoniaceae plastomes, including those of Begonia’s sister group, Hillebrandia, a monotypic genus endemic to Hawai‘i, and 43 species representing 42 sections of Begonia, were assembled. Our results reveal that Begoniaceae plastome size ranges from 167,123 to 170,852 bp, displaying the typical quadripartite structure. Structures of most Begoniaceae plastomes are highly conserved but differ from the plastomes of the majority of angiosperms in having a unique inverted repeat (IR) expansion, from IRa to large single copy (LSC), resulting from a duplicated fragment of the trnH–GUG gene to the trnR–UCU gene. Additionally, comparison between plastomes of Hillebrandia and Begonia shows that the former genus has fewer simple sequence repeats than most Begonia species analysed, suggesting that species of Begonia have more repetitive and dynamic plastomes than those of its sister genus. We also identified six highly variable regions suitable for phylogenetic analysis and as potential DNA barcodes for species identification. Our robust hypothesis of plastome phylogenomic relationships provides new insights into infrageneric classification and highlights potential classification issues in Begonia.
Downloads
Published
Issue
Section
License
Copyright (c) 2022 Y.-H. Tseng, C.L. Hsieh, L. Campos-Domínguez, A.Q. Hu, C.C. Chang, Y. T. Hsu, C.A. Kidner, M. Hughes, P.W. Moonlight, C.H. Hung, Y.C. Wang, Y.T. Wang, S.H. Liu, D. Girmansyah, K.-F. Chung

This work is licensed under a Creative Commons Attribution 4.0 International License.
Please read our Open Access, Copyright and Permissions policies for more information.

